| |
||
|
Proteomic Analysis of Arabidopsis
thaliana in Microgravity The function and appearance of cells (and the organism that the cells compose) are determined by the specific proteins the cells produce. The overall goal of this project is to determine which proteins plants produce, degrade, and modify in response to growth in space. To do this, we're going to send some Arabidopsis seedlings to grow on the international space station, while identical control plants are kept on earth for comparison. We'll then use Proteomics to compare the total protein content of each set of plants. There are several potential benefits from this research. Extended, manned space exploration will require plant growth for both food and oxygen. Understanding how plants handle the stresses of space, not only on a physioilogical level, but also a molecular one, will be critical. In addition, this analysis will reveal candidate genes that we can use to better understand how plants respond to the constant force of gravity here on earth. This has potential to inform agricultural practices, as well as provide specific targets for genetic engineering.
|
||
|
Click here for NASA project photos (opens a new tab) |
||
| |
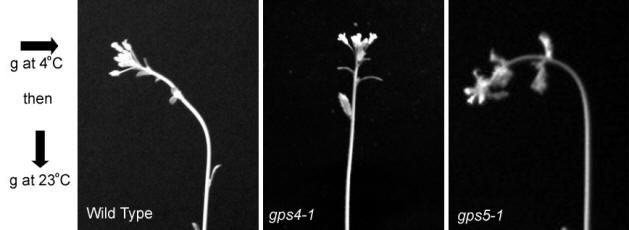
GravitropisM To gain a unique perspective on the well-studied process of gravitropism, we focus on the Gravity Persistent Signal (GPS) response as a means to dissect cell signaling in Arabidopsis (Fukaki et al., 1996). At cold temperatures (4°C), Arabidopsis inflorescence stems do not respond to gravity due to an inhibition of auxin transport (Nadella et al., 2006). However, when a plant is subjected to a 90° gravistimulation at 4°C, and subsequently returned to room temperature in a vertical orientation, the inflorescence stem displays a transient bending in the direction that would have been “up” when it was oriented horizontally in the cold. The bottom line is that a plant ends up bending to the side at room temperature after a gravistimulation in the cold. This is referred to as the Gravity Persistent Signal (GPS) response. It is thought to be caused by a buildup of signaling proteins and secondary messengers during the cold treatment that have no outlet without the ability to transport auxin. Previous work suggests that the GPS process is ideal for analyzing the gravitropic pathway in a simplified system, allowing for the discovery of signaling components that may otherwise have undetectable mutant phenotypes due to redundant function (Wyatt et al., 2002). Currently, the lab is focused on two gravity persistent signal (gps) mutants in Arabidopsis that display an aberrant response to gravity after a cold treatment. One of these, gps4, displays no gravitropism after a GPS treatment. Although its molecular identity is known, the exact function of GPS4 remains a mystery. The gps5 mutant displays enhanced gravitropism in the root, hypocotyl and inflorescence stem after a 90° gravitropic stimulus. Furthermore, gps5 has drastically increased root and hypocotyl growth. Currently, research is focused on identifying the molecular target of GPS5, and determining its role in plant growth and development. This project has been supported by the Summer Research Fellowship and the Undergraduate Research and Creative Activities awards at SIUE.
|
|
| |
||

| |
||
| |
Chlorophyll Synthesis  ggps1-1 |
|
| |
||
 Time
course of ggps1-1 growth in cool and warm
conditions. Figure by Kelsey Kropp.
Time
course of ggps1-1 growth in cool and warm
conditions. Figure by Kelsey Kropp.
Publications
Ruppel, NJ, Kropp, KN, Davis, PA, Martin, AE, Luesse, DR, and Hangarter, RP. (2013) Mutations in GERANYLGERANYL DIPHOSPHATE SYNTHASE 1 affect chloroplast development in Arabidopsis thaliana. American Journal of Botany 100(10): 2074-2084.
Luesse, DR, Schenck, CA, Berner, BK, Justus, B, Wyatt, SE (2010) GPS4 is allelic to ARL2: Implications for gravitropic signal transduction. Gravitational and Space Biology 23: 95-96.
Luesse, DR, DeBlasio, SL, Hangarter RP (2010) Integration of phot1, phot2, and PhyB signalling in light-induced chloroplast movements. Journal of Experimental Botany. 61: 4387-4397.
Luesse, DR, DeBlasio, SL, Hangarter RP (2006) Plastid Movement Impaired 2 (PMI2 a new gene required for normal blue-light-induced chloroplast movements in Arabidopsis thaliana. Plant Physiology. 141: 1328-1337.
DeBlasio SL, Luesse DR, Hangarter RP (2005) A plant-specific protein essential for blue-light-induced chloroplast movements. Plant Physiology. 139: 101-114
DeBlasio SL, Mullen JL, Luesse DR, Hangarter RP (2003) Phytochrome modulation of blue light-induced chloroplast movements in Arabidopsis. Plant Physiology. 133: 1471-1479
Stowe-Evans EL, Luesse DR, Liscum E (2001) The enhancement of phototropin-induced phototropic curvature in arabidopsis occurs via a photoreversible phytochrome A-dependent modulation of auxin responsiveness. Plant Physiology 126: 826-834
Harper RM, Stowe-Evans EL, Luesse DR, Muto H, Tatematsu K, Watahiki MK, Yamamoto K, Liscum E (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12: 757-770
|
Dr. Darron R. Luesse Office:
1125 Science Lab Building West email: dluesse@siue.edu
Address: Box 1651 Southern Illinois University Edwardsville, IL 62025
|

|